Varbina Ivanova / Jordi Juárez (UB) — Multiple‑copy association studies (MAS) as a tool for Computer‑Aided Drug Design
Don't miss any Success Story following us on X and LinkedIn!
@RES_HPC RES - Red Española de Supercomputación @res-icts.bsky.social
Check this Success Story at our LinkedIn: Multiple‑copy association studies (MAS) as a tool for Computer‑Aided Drug Design
💡 A RES Success Story about accelerating protein-ligand binding mode prediction without prior binding site knowledge 💡
📋 "Multiple‑copy association studies (MAS) as a tool for Computer‑Aided Drug Design" developed by Varbina Ivanova and Jordi Juárez Jiménez from Universitat de Barcelona
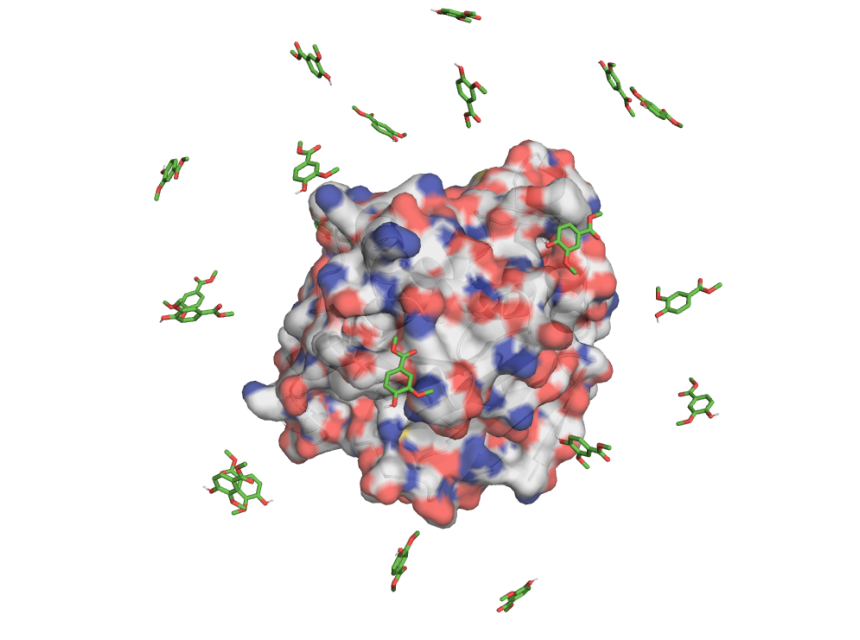
Predicting how small molecules bind to proteins is critical for drug discovery, but challenging when there isn't structural data available. To address this, the team developed and validated the MAS workflow, which combines high‑ligand‑concentration Molecular Dynamics (MD) simulations with clustering and stability analysis to identify probable protein-ligand binding modes without prior information about the binding site.
🖥 Thanks to RES supercomputer #MareNostrum5 from Barcelona Supercomputing Center, the team performed large-scale GPU-accelerated molecular dynamics simulations of multiple protein–ligand systems to develop and validate the MAS workflow. These simulations involved placing multiple ligand copies around the protein of interest and running unbiased MD replicas to explore the ligand binding across the entire protein surface.
The workflow showed excellent performance, successfully recovering the correct binding site in 22 out of the 23 test cases and the crystallographic binding mode in 16 cases, without any prior knowledge of the ligand's binding site. This demonstrates MAS as a powerful and scalable strategy to accelerate computer-aided drug discovery and guide hit-to-lead optimization when structural data is limited.
🎥 The video illustrates an example from the MAS validation, where multiple ligand copies explore the protein surface during a high-ligand-concentration MD simulation and one ligand successfully converges to the experimentally determined binding mode.
📋 "Multiple‑copy association studies (MAS) as a tool for Computer‑Aided Drug Design" developed by Varbina Ivanova and Jordi Juárez Jiménez from Universitat de Barcelona
Predicting how small molecules bind to proteins is critical for drug discovery, but challenging when there isn't structural data available. To address this, the team developed and validated the MAS workflow, which combines high‑ligand‑concentration Molecular Dynamics (MD) simulations with clustering and stability analysis to identify probable protein-ligand binding modes without prior information about the binding site.
🖥 Thanks to RES supercomputer #MareNostrum5 from Barcelona Supercomputing Center, the team performed large-scale GPU-accelerated molecular dynamics simulations of multiple protein–ligand systems to develop and validate the MAS workflow. These simulations involved placing multiple ligand copies around the protein of interest and running unbiased MD replicas to explore the ligand binding across the entire protein surface.
The workflow showed excellent performance, successfully recovering the correct binding site in 22 out of the 23 test cases and the crystallographic binding mode in 16 cases, without any prior knowledge of the ligand's binding site. This demonstrates MAS as a powerful and scalable strategy to accelerate computer-aided drug discovery and guide hit-to-lead optimization when structural data is limited.
🎥 The video illustrates an example from the MAS validation, where multiple ligand copies explore the protein surface during a high-ligand-concentration MD simulation and one ligand successfully converges to the experimentally determined binding mode.