Allan Mackie / Merve Gözdenur / Clara Salueña (URV) — Coarse-grain ssDNA simulations with Dissipative Particle Dynamics using the MARTINI 'Lego' approach
Don't miss any Success Story following us on X and LinkedIn!
@RES_HPC RES - Red Española de Supercomputación @res-icts.bsky.social
Check this Success Story at our LinkedIn: Coarse-grain ssDNA simulations with Dissipative Particle Dynamics using the MARTINI 'Lego' approach
💡 A RES success story about coarse-grain simulations of membraneless organelles
📋"Coarse-grain ssDNA simulations with Dissipative Particle Dynamics using the MARTINI 'Lego' approach" led by Allan D. Mackie, Clara Salueña and Merve Gözdenur Demirbek from ETSEQ at Universitat Rovira i Virgili
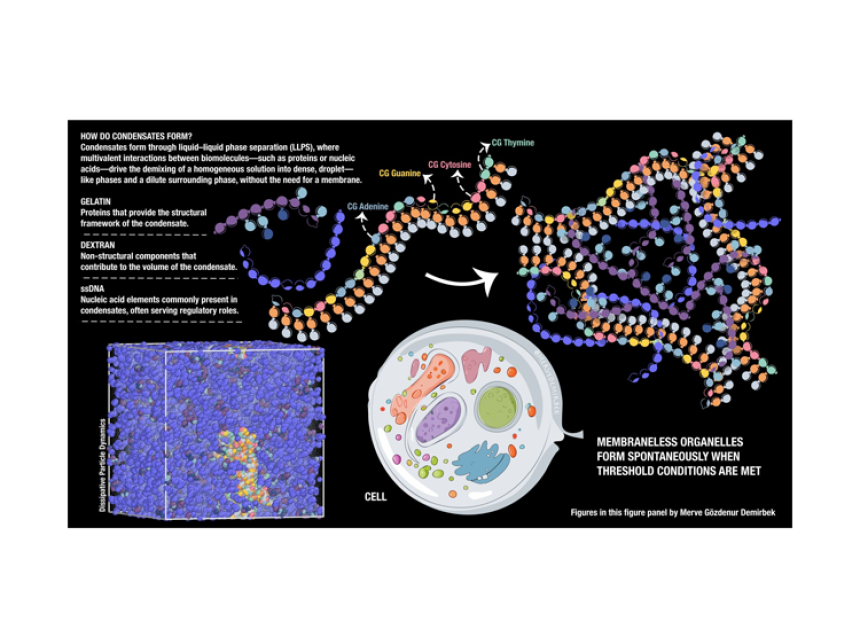
The self-organization of biomolecules into functional structures is a fundamental process in biology. One example of this phenomenon is the formation of membraneless organelles (MLOs) through liquid-liquid phase separation (LLPS), driven by multivalent interactions between proteins, nucleic acids, and polysaccharides. These condensates, such as nucleoli and stress granules, play essential roles in cellular processes without requiring lipid membranes.
🖥️ Thanks to RES supercomputer #MareNostrum5 from Barcelona Supercomputing Center, the team was able to perform large-scale, time-resolved simulations of an aqueous mixture of ssDNA, dextran, and gelatin in various conditions.
The team used Dissipative Particle Dynamics (DPD) simulations to explore the formation and behaviour of MLOs under biologically relevant conditions. Particularly, they modeled coarse-grained single-stranded DNA (ssDNA), gelatin (representing a protein component), and dextran (representing a polysaccharide) to replicate the complex nature of MLOs. The main objective of this project is to create a flexible and scalable toolkit with MARTINI3 within a DPD framework to study complex biological phenomena and phase behaviour at mesoscale resolution.
📸 This image illustrates the self-assembly of biomolecules into membraneless organelles through liquid-liquid phase separation, as modeled using coarse-grained simulations.
📋"Coarse-grain ssDNA simulations with Dissipative Particle Dynamics using the MARTINI 'Lego' approach" led by Allan D. Mackie, Clara Salueña and Merve Gözdenur Demirbek from ETSEQ at Universitat Rovira i Virgili
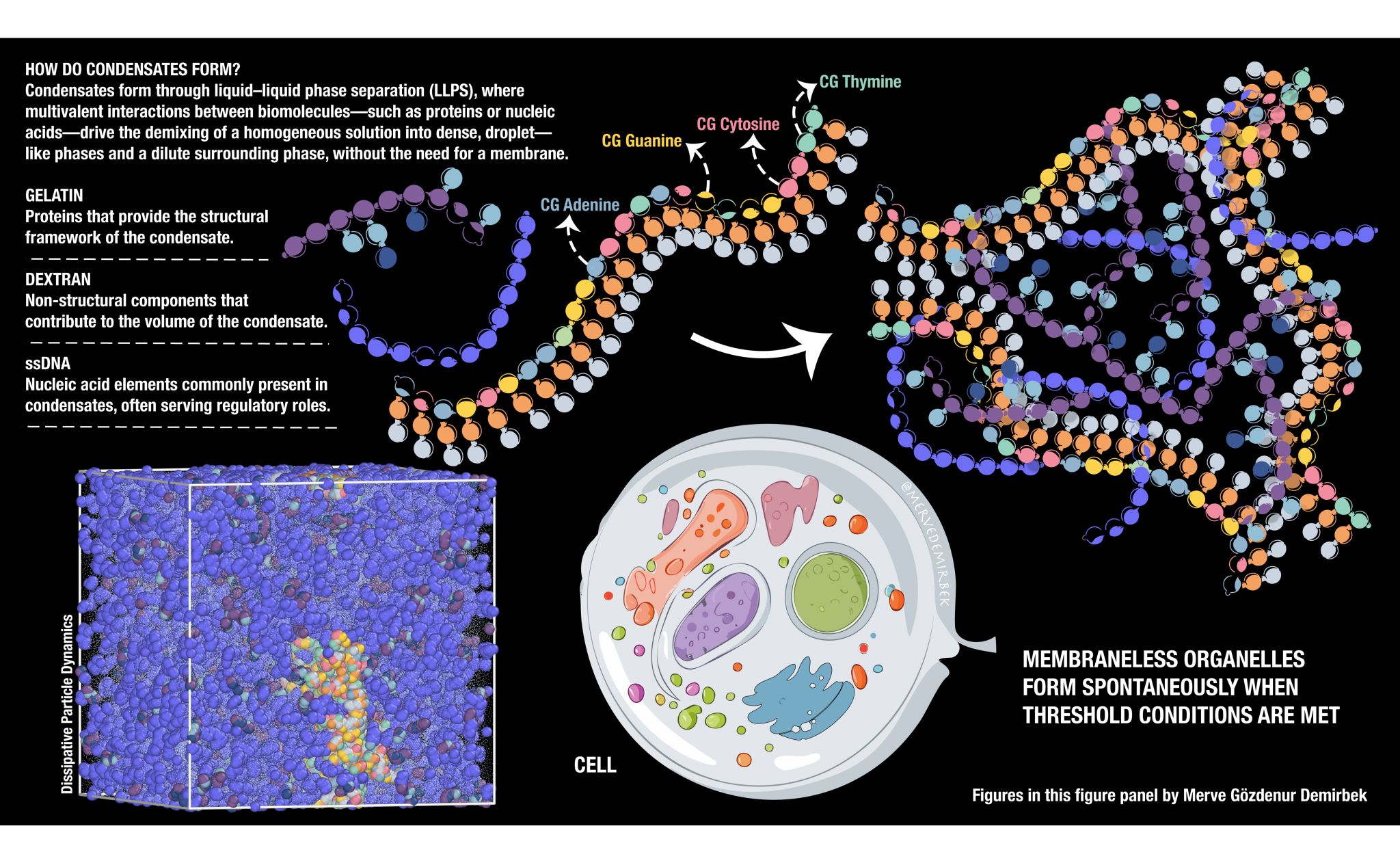
The self-organization of biomolecules into functional structures is a fundamental process in biology. One example of this phenomenon is the formation of membraneless organelles (MLOs) through liquid-liquid phase separation (LLPS), driven by multivalent interactions between proteins, nucleic acids, and polysaccharides. These condensates, such as nucleoli and stress granules, play essential roles in cellular processes without requiring lipid membranes.
🖥️ Thanks to RES supercomputer #MareNostrum5 from Barcelona Supercomputing Center, the team was able to perform large-scale, time-resolved simulations of an aqueous mixture of ssDNA, dextran, and gelatin in various conditions.
The team used Dissipative Particle Dynamics (DPD) simulations to explore the formation and behaviour of MLOs under biologically relevant conditions. Particularly, they modeled coarse-grained single-stranded DNA (ssDNA), gelatin (representing a protein component), and dextran (representing a polysaccharide) to replicate the complex nature of MLOs. The main objective of this project is to create a flexible and scalable toolkit with MARTINI3 within a DPD framework to study complex biological phenomena and phase behaviour at mesoscale resolution.
📸 This image illustrates the self-assembly of biomolecules into membraneless organelles through liquid-liquid phase separation, as modeled using coarse-grained simulations.